 |
|
News and Publications
Synthesis of Multiresolution Structural Data
W. Wriggers, P. Chacón, C.L. Brooks III, R.A. Milligan, E. Kim*
* University of California, Los Angeles, CA
Future advances in modern biology and medicine will depend on an
understanding of fundamental cellular processes, most of which involve
the actions and interactions of large biomolecular aggregates.
Three-dimensional structures and image reconstructions of aggregates,
involving hundreds of thousands to millions of atoms, are now routinely
determined by using x-ray crystallography and electron microscopy. To
further our understanding of cellular function, we must be able to
characterize the architecture and functionally relevant motions of
these cellular machines at all levels of resolution.
 The
Computational Structural Biology program is active both in developing
computational tools for multiresolution docking and modeling and in
applying this technology to the study of large-scale molecular
machines, including molecular motors (myosin and kinesin) and their
tracks (actin and microtubules), virus capsids, and the ribosome. We
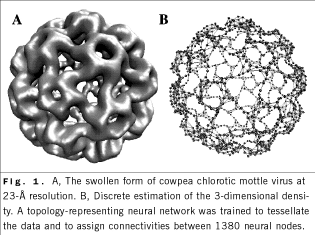
use topology-representing neural networks for a coarse estimation of
density maps and for determining suitable landmarks for the
registration of multiresolution data (Fig. 1). In the past year, we
showed the usefulness of this approach in a variety of docking
applications and developed the program package Situs to provide a
powerful method for the localization of biomolecular subunits in
low-resolution data from electron microscopy and small-angle x-ray
scattering. Using neural networks to examine discrete parts of the
search space reduced the search times for optimum alignment of
3-dimensional data from hours to seconds. The
Computational Structural Biology program is active both in developing
computational tools for multiresolution docking and modeling and in
applying this technology to the study of large-scale molecular
machines, including molecular motors (myosin and kinesin) and their
tracks (actin and microtubules), virus capsids, and the ribosome. We
use topology-representing neural networks for a coarse estimation of
density maps and for determining suitable landmarks for the
registration of multiresolution data (Fig. 1). In the past year, we
showed the usefulness of this approach in a variety of docking
applications and developed the program package Situs to provide a
powerful method for the localization of biomolecular subunits in
low-resolution data from electron microscopy and small-angle x-ray
scattering. Using neural networks to examine discrete parts of the
search space reduced the search times for optimum alignment of
3-dimensional data from hours to seconds.
The rigid-body docking technique also laid the groundwork for
the development of a flexible docking technique that brings deviating
features of multiresolution structures into register. We devised a
novel flexible docking procedure based on artificial neural networks
and molecular dynamics simulation that improves the agreement between
data sets in the presence of "induced fit" conformational changes. This
approach was applied to the flexible docking of the ribosomal
elongation factor EF-G into electron microscopy density maps.
Finally, we investigated how to include geometric constraints
from a variety of biophysical sources in the modeling: electron
microscopy, small-angle x-ray scattering, fluorescence spectroscopy,
mutagenesis, and biochemical labeling and footprinting. For example, we
refined the atomic model of actin filaments by using cysteine
cross-links that were introduced experimentally and that provide
stringent constraints for the arrangement of actin subunits and their
contact interface. To investigate the effect of H73, conserved in all
actin species, on protein structure, we complemented mutagenesis
experiments with simulated annealing simulations of the mutants that
revealed an effect of the electrostatic charge at this position on the
forming of salt bridges near the active site.
PUBLICATIONS
Kim, E., Wriggers, W., Phillips, M., Kokabi, K., Rubenstein, P., Reisler, E. Cross-linking constraints on F-actin structure. J. Mol. Biol. 299:421, 2000.
Wriggers, W., Agrawal, R.K., Drew, D.L., McCammon, J.A., Frank, J.
Domain movements of EF-G bound to the 70S ribosome: Insights from a
hand-shaking between multi-resolution structures. Biophys. J. 79:1670,
2000.
Wriggers, W., Milligan, R.A., McCammon, J.A. Situs: A package
for docking crystal structures into low-resolution maps from electron
microscopy. J. Struct. Biol. 125:185, 1999.
Xing, J., Wriggers, W., Jefferson, G.M., Stein, R., Cheung, H.C., Rosenfeld, S.S. Kinesin has three nucleotide-dependent conformations: Implications for strain-dependent release. J. Biol. Chem., in press.
Yao, X., Grade, S., Wriggers, W., Rubenstein, P. His73, often methylated, is an important structural determinant for actin: A mutagenic analysis of His73 of yeast actin. J. Biol. Chem. 274:37443, 1999.
|
|



