
![]()



![]()

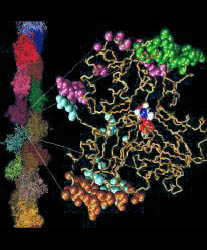
Fig. 1 (click to enlarge): Actin monomers before and after
counterion binding. Top: In the absence of calcium ions, the protein is
negatively charged. Bottom: After binding of five counterions at
predicted sites, the protein is neutralized. The isocontours (red) at
-3 kcal/mol were calculated with the program Grasp at 0 ionic strength.
Our research is concerned, in particular, with the divalent-cation-driven polymerization of actin (described elsewhere). The aggregation requires the binding of divalent cations to five low-affinity sites (Kd ~15mM), neutralizing the monomers (charge at pH 7: -10 e). The location of these sites in the actin crystal structure is unknown. Brownian dynamics studies of actin-binding proteins and actin are under way in several research groups. As outlined above, these studies require the prediction of actin's divalent cation binding sites.
We have initiated searches for actin's five divalent cation binding sites and computed the free energy of binding at 10,000 equally-spaced test sites on actin's ion-accessible surface using the program UHBD (Fig. 2). These preliminary calculations will be followed by more extensive studies including side chain flexibility, ion cooperativity, and a ranking of the sites by the free energy of binding.

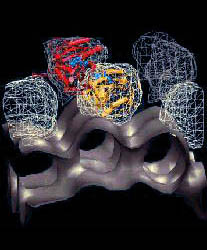
Fig. 2 (click to enlarge): Free energy of calcium binding to the
actin monomer surface. Red: -5 kcal/mol; green: 0, blue; +5 kcal/mol.
Finally, we collaborate with the laboratory of Gerard Wong to determine directly the binding of counter-ion clouds to actin filament bundles by small-angle x-ray scattering (see reference below).
![]()
![]()