RNA Polymerase Flexing and Animations
A flexible alignment tool was recently added to our 'Situs'
docking package that is based on 3D 'motion capture' technology used in
the entertainment industry and in biomechanics. We have used this tool
in collaboration with Seth Darst
(Rockefeller University), whose laboratory determined the structure of
Escherichia coli core RNA polymerase (RNAP) by cryo-electron microscopy
and image processing of helical crystals to a resolution of 15Å.
Because of the high sequence conservation between the core RNAP
subunits, we were able to interpret the E. coli structure in relation
to the high-resolution X-ray structure of Thermus aquaticus core RNAP.
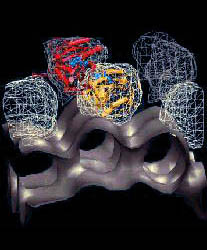
A very large conformational change of the T. aquaticus RNAP X-ray
structure, corresponding to opening of the main DNA/RNA channel by
nearly 25Å, was required to fit the E. coli map (see Figure).
This finding reveals, at least partially, the range of conformational
flexibility of the RNAP, which is likely to have functional
implications for the initiation of transcription, where the DNA
template must be loaded into the channel.

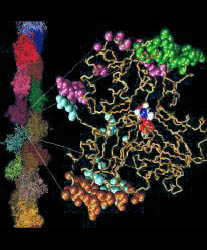
Figure: Direct space flexible fitting with skeletons. This
overview illustrates the skeleton-based modeling steps that were
employed in the recent flexible refinement of the T. aquaticus crystal
structure (upper right) of RNA polymerase against EM data (upper left)
from the laboratory of Seth Darst. The flexed model (bottom) is
compared to the EM data (bottom right). By freezing inessential degrees
of freedom, skeletons of connected landmarks significantly reduce the
effect of noise and thereby improve the stereochemical quality of the
fitted structures relative to unconstrained alignments.
Movies:
These
animations created by Seth Darst demonstrate the conformational change
induced by the transition from the X-ray structure to the EM envelope.
The color codes for the actual displacement, where warm colors move
more than cold colors.
References:
- Natacha Opalka, Mark Chlenov, Pablo Chacón, William J. Rice, Willy Wriggers, and Seth Darst. Structure and Function of the Transcription Elongation Factor GreB Bound to Bacterial RNA Polymerase. Cell, 2003, Vol. 114, pp. 335-345.
[Abstract]
[Article]
[Cell Introduction]
- Seth A. Darst, Natacha Opalka, Pablo Chacón, Andrey Polyakov,
Catherine Richter, Gongyi Zhang, and Willy Wriggers. Conformational
Flexibility of Bacterial RNA Polymerase. PNAS, 2002, Vol. 99, pp. 4296-4301.
[Abstract]
[Article]
[Structure Paper Alert] [Predicted Structure and Movies]
- Willy Wriggers and Pablo Chacón. Modeling Tricks and
Fitting Techniques for Multi-Resolution Structures. Structure,
2001, Vol. 9, pp. 779-788. [1st Paragraph]
[Article]



![]()



![]()

![]()
![]()