Actin: Simulations and Animations
Actin
filaments are dynamic polymers whose ATP-driven assembly in the cell
cytoplasm drives shape changes, cell locomotion and chemotactic
migration. Actin filaments also participate in muscle contraction. The
structure of the filament is not known at atomic resolution, but
several models were produced in the laboratory of Ken Holmes (MPI for
medical research, Heidelberg, Germany) by refinement against X-ray
fiber diffraction data:

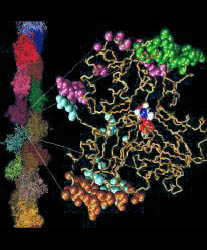
Fig. 1 (click to enlarge): Lorenz model of F-actin. A single
G-actin monomer with inter-actin contact surfaces is shown on the
right, the entire F-actin on the left. This Figure, created with the
graphics program VMD,
is actually quite popular and has been downloaded by numerous
researchers and printed in two textbooks. The figure is copyrighted.

1. Actin's "Back Door"
The
mechanical properties of the filaments can be altered by actin-binding
agents such as the toxin phalloidin from the mushroom "Amanita
phalloides". Phalloidin binding to actin has been shown to delay the
release of inorganic phosphate after ATP hydrolysis (Dancker &
Hess, Biochim. Biophys. Acta, 1990, 1035:197). Conversely, no effect of
inorganic phosphate on the phalloidin binding kinetics has been
observed (De La Cruz & Pollard, Biochemistry, 1994, 33:14378). Our
research goal was to resolve this seemingly paradoxical situation by
identifying the dissociation pathway of the phosphate.
The
simulation of monomeric actin required the development of a proper
force field for ATP and ADP nucleotides (parameters available by download).
The major discovery was the diffusion of water molecules into the
buried nucleotide binding site along two distinct pathways (Fig. 2). Of
special interest is the "back door" diffusion pathway which we believe
to be relevant to the dissociation of the phosphate after hydrolysis.
Our hypothesis for phosphate release would explain the seemingly
paradoxical kinetics of actin-phalloidin interaction. Phalloidin, whose
position in the F-actin filament structure is known, would in our model
block the exit of the back door pathway and, thus, delay the release of
the phosphate.

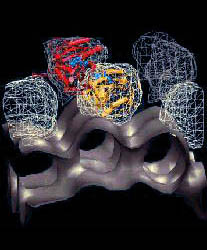
Fig. 2 (click to enlarge): Diffusion of water molecules into the nucleotide binding site of actin [visualized with the program VMD (side view)].
ATP and the Mg++ ion are represented as white spheres in the center of
the protein, which is rendered as a transparent ribbon. The color of
the traces of the water oxygens codes for different simulation
trajectories. The Figure reveals two water diffusion pathways with the
"back door" pathway on the right.

2. Actin Phosphate Release - The Movie
In
order to substantiate our back door hypothesis of actin phosphate
dissociation we studied further the dissociation of the nucleotide and
the role of the proposed back door pathway for actin's enzymatic
activity. To reveal the microscopic processes underlying the release of
the inorganic phosphate, we explored possible release pathways in steered molecular dynamics
simulations by measuring adhesion forces when pulling the substrate out
of its binding pocket. The resulting structural prediction of the
dissociation mechanism has important implications also for other
putative back door enzymes such as the ATP-driven motor protein myosin
(Yount et al., Biophys J., 1995, 68:44s).
Animations (total length 4 minutes):
Based
on these studies, we proposed that the imidazole moiety of histidine
73, which is methylated in most actins, interacts with the dissociating
phosphate and that electrostatic stabilization retards its release from
actin following ATP hydrolysis. To investigate the effect of H73
further, mutagenesis experiments (substitution with R,K,A,E,Q) were
performed in the laboratory of Peter Rubenstein.
The observed trends in these experiments are consistent with results
obtained by simulated annealing simulations of the mutant actins. Two
publications summarizing the latest experimental and simulation results
were published in JCB (see below).

3. Actin Filament Severing by Cofilin

Fig. 3 (click
to enlarge): Cover figure of the Oct. 9, 1998, issue of J. Molec. Biol.
Shown on the left are the nonbonded interaction energies arising in the
complex of cofilin (top) with actin (bottom). Shown on the right are
the contact surfaces in the complex, colored by atom type.
An understanding of the actin-depolymerizing function attributed to members of the
ADF/cofilin/destrin superfamily requires a structural model of these proteins in complex with
actin. As a step toward defining actin-cofilin interactions, the complex of yeast cofilin with
monomeric actin was predicted by molecular dynamics simulation in collaboration with Jay X. Tang and Paul A. Janmey. The predicted complex exhibits
strong interactions between the N-termini of actin and cofilin, mediated by a salt bridge of
cofilin Arg3 with actin Asp1. The forming of this salt bridge could be prevented by the phosphorylation of cofilin Ser4 which is
believed to inhibit cofilin depolymerization activity. Recent mutagenesis studies, crosslinking
experiments and peptide binding studies are consistent with the predicted model of the
actin-cofilin complex. The structural homology between cofilin and gelsolin segment-1 binding
to actin was confirmed experimentally by two types of competitive binding assays.

Fig. 4 (click to enlarge): Filament severing by steric
exclusion. When aligned to the actin filament in the monomer binding
mode, cofilin exhibits steric clashes with actin's subdomain 2 (black).
The model is consistent with a conformational transition from the
filamentous to the monomeric cofilin binding mode that weakens the
interactions of adjacent actin monomers in the filament.
More
recently, we modelled the atomic structure of the cofilin-F-actin
complex based on electron microscopy data from the laboratory of Edward H. Egelman:

Fig. 5 (click to enlarge): Cover figure of the April 2, 2001
issue of Journal of Cell Biology. The image shows the atomic model
(left) and an electron microscopy map (right) of doubly ADF-decorated
actin filaments.

4. Cross-Links and Actin Filament Structure
We
refined the model of F-actin further by modeling of cysteine
cross-linking constraints on the structure. The cross-links are
introduced experimentally in the laboratory of Emil Reisler
and provide stringent constraints for the arrangement of actin subunits
and their contact interface. The DNase I binding loop (residues 38-52),
the hydrophobic plug (residues 262-274), and the C-terminus region are
among the structural elements of monomeric (G-) actin that were
proposed to form the intermonomer interface in F-actin. Cysteines were
introduced into yeast actin either at residue 41 or 265. These
mutations allowed for specific cross-linking of F-actin between C41 and
C265, C265 and C374, and C41 and C265. The calculated cross-linked
structures showed a better fit to the Holmes than to the refined Lorenz
model of F-actin. It is predicted on the basis of such calculations
that image reconstruction of electron
micrographs of disulfide cross-linked C41-C374 F-actin should provide a
conclusive test of these two similar models of F-actin structure.

Fig.
6 (click to enlarge): Multi-resolution rendering of conformational
changes induced by the 41-374 disulfide bridge. (a) The Holmes model of
F-actin after refinement. Individual monomers
are shown as colored backbone traces, Ca2+-ADP as black spheres. The
41-374 cross-
bridges are shown in red. (b) Low-resolution map of the original Holmes
model, shown
as an isocontour at half-maximal density. The spatial resolution of the
map (full width
half-maximum of the Gaussian kernel) is 13.6A (c) Low-resolution map of
the refined
model shown as a wire-mesh, otherwise rendered as in (b). In addition,
the difference
density map relative to (b) is visualized by a red isocontour at -10%
of the maximum
density value. Positive difference densities are below the +10% density
threshold. (d) The
refined Lorenz model with the cross-linked sites (red). (e)
Low-resolution map of the
original Lorenz model, shown as an isocontour at half-maximal density.
(f) Low-resolution map of the Lorenz model in the presence of the
41-374 cross-bridge shown as
a wire-mesh. The difference density map relative to (e) is visualized
by isocontours
at -10% (red) and + 10% (blue) of the maximum density value.

References
- Xiaoyi Yao, Vinh Nguyen, Willy Wriggers, and Peter Rubenstein.
Regulation of Yeast Actin Behavior by Interaction of Charged Residues Across
the Interdomain Cleft. J. Biol. Chem., 2002, Vol. 25, pp. 22875-22882.
[Abstract]
[Article] [Predicted Structures]
- Vitold E. Galkin, Albina Orlova, Natalya Lukoyanova, Willy Wriggers,
and Edward H. Egelman. ADF Stabilizes an Existing State of F-actin
and Can Change the Tilt of F-actin Subunits. J. Cell Biol., 2001,
Vol. 153, pp. 75-86 [Abstract]
[Article]
[Predicted Structure]
- Eldar Kim, Willy Wriggers, Martin Phillips, Kevin Kokabi, Peter
Rubenstein, and Emil Reisler. Cross-Linking Constraints on F-actin
Structure. J. Molec. Biol., 2000, Vol. 299, pp. 421-429.
[Abstract]
[Article]
[Predicted Structures]
- Xiaoyi Yao, Stephanie Grade, Willy Wriggers, and Peter Rubenstein.
His73, Often Methylated, Is an Important Structural Determinant for Actin
- A Mutagenic Analysis of H73 of Yeast Actin. J. Biol. Chem., 1999,
Vol. 274, pp. 37443-37449. [Abstract]
[Article] [Predicted Structures]
- Willy Wriggers and Klaus Schulten. Investigating a Back Door
Mechanism of Actin Phosphate Release by Steered Molecular Dynamics.
Proteins: Structure, Function, and Genetics, 1999, Vol. 35, pp. 262-273.
[Abstract]
[Article]
- Sergei Izrailev, Sergey Stepaniants, Barry Isralewitz, Dorina Kosztin,
Hui Lu, Ferenc Molnar, Willy Wriggers, and Klaus Schulten. Steered Molecular
Dynamics. Chapter in "Computational Molecular Dynamics: Challenges, Methods,
Ideas", Springer Verlag, 1999
[Abstract]
[Article]
- Willy Wriggers, Jay X. Tang, Toshifumi Azuma, Peter Marks, and
Paul A. Janmey. Cofilin and Gelsolin Segment-1: Molecular Dynamics Simulation
and Biochemical Analysis Predict a Similar Actin Binding Mode. J. Molecular
Biology, 1998, Vol. 282, pp. 921-932.
[Abstract]
[Article]
[Predicted Structure]
- Willy Wriggers and Klaus Schulten. Stability and Dynamics
of G-actin: Back Door Water Diffusion and Behavior of a Subdomain 3/4 Loop.
Biophysical Journal 1997, Vol. 73, pp. 624-639.
[Abstract]
[Article]
[Nucleotide Parameters]
- Thomas E. Angelini, Hongjun Liang, Willy Wriggers and Gerard Wong. Like-charge attraction between polyelectrolytes induced by counterion charge density waves. PNAS, 2003, Vol. 100, pp. 8634-8637.
[Abstract]
[Article] [Software]


![]()



![]()

![]()

![]()
![]()



![]()
![]()
![]()